Solution | Medical Entity Extraction Assistant#
Overview#
Business case#
An estimated 80% of electronic health/medical records (EHR/EMR) data is unstructured. This data comes from medical imaging (such as X-ray, CT, MRI) and free-form text from healthcare providers (such as clinical notes, nurse admission notes, doctors’ discharge summaries). The volume is increasing due to digital and telehealth engagement, generating new data such as patient-reported outcomes and transcripts. This deluge has become an incredible challenge for medical reviewers and coders, creating a critical resource gap that prevents them from processing incoming medical texts.
This challenge is particularly acute with the increasing demands of value-based care (VBC) programs. Written notes often capture rich information about the severity of patients’ conditions, interventions, clinical recommendations, and even socioeconomic status. In contrast, structured medical codes from claims or EHR data—often designed for billing and transactional purposes—fail to represent a complete view of a patient’s health.
Although clinical and administrative staff can access more comprehensive patient information by reviewing these notes, the process remains highly manual and time-consuming.
Recent advances in Generative AI have shown that large language models (LLMs) can perform numerous tasks in a medical context, including summarizing doctor-patient transcripts, answering medical licensing exams, and supporting medical consultations. However, any automation or AI used to accelerate this process must incorporate human-in-the-loop verification and validation. This is particularly critical because foundation models often perform suboptimally when extracting medical codes from text.
This Solution proposes a Generative AI (GenAI) framework that extracts critical medical concepts and maps them to standardized vocabularies. It includes a human-in-the-loop interface to facilitate expert validation of the GenAI-assigned codes. It also offers a quick dashboard with metrics to examine the GenAI pipeline’s performance on code generation.
Key beneficiaries include:
Medical coders
Clinicians
Epidemiologists and health outcomes researchers
Installation#
This Solution is currently in a private preview phase. If you’re interested in accessing this Solution, please reach out to your Dataiku account manager or use the general Contact Us form.
Technical requirements#
To use this Solution, you must meet the following requirements:
Have access to a Dataiku 14.2+* instance.
A Python 3.9 code environment named
solution_med-entity-extract-assist, with the following required packages:
dash==3.1.1
dash-extensions==1.0.20
dash-bootstrap-components==2.0.3
An internal code environment for retrieval augmented generation. Please see the reference documentation on Initial setup.
The VisualEdit Plug-in V2.0.10.
Data requirements#
Data preparation and feature engineering should be completed in a separate project to match the expected input dataset format. Adhering to the expected data model ensures reliable and interpretable results.
Input datasets format#
Clinical notes
Required schema for the input dataset:
column name |
description |
|---|---|
patient_id |
Patient’s unique identifier |
note_id |
Clinical note’s unique identifier |
sex |
Patient’s sex |
age |
Patient’s age |
note |
Clinical note full text |
Workflow overview#

The project has the following high-level steps:
Extracts key clinical events and associated medical concepts
Assigns standardized billing codes (for example ICD10CM and ICD10PCS) to each medical concept
Interactive webapps for users to review, edit, and verify the model-assigned codes
A model dashboard to assess LLM performance on medical code generation.
Walkthrough#
Solution set-up via Dataiku app#
To begin, the Solution must be set up via the Dataiku app. This process ingests the clinical note file and prepares the pipeline to generate medical codes. The Dataiku app is available on the homepage of Dataiku under the Applications section.
To begin, you’ll need your own instance of the Dataiku app associated with this Solution.
From the waffle (
) menu of the Design node’s top navigation bar, select Dataiku Apps.
Search for and select the application with the name of this Solution.
Then, click Create App Instance.
This will create a new instance of the parent project, which you can configure to suit your specific needs. It will help you connect to your data regardless of the connection type and configure the Flow based on your parameters. You can make as many instances as needed (for example, if you want to apply this Solution to other data).
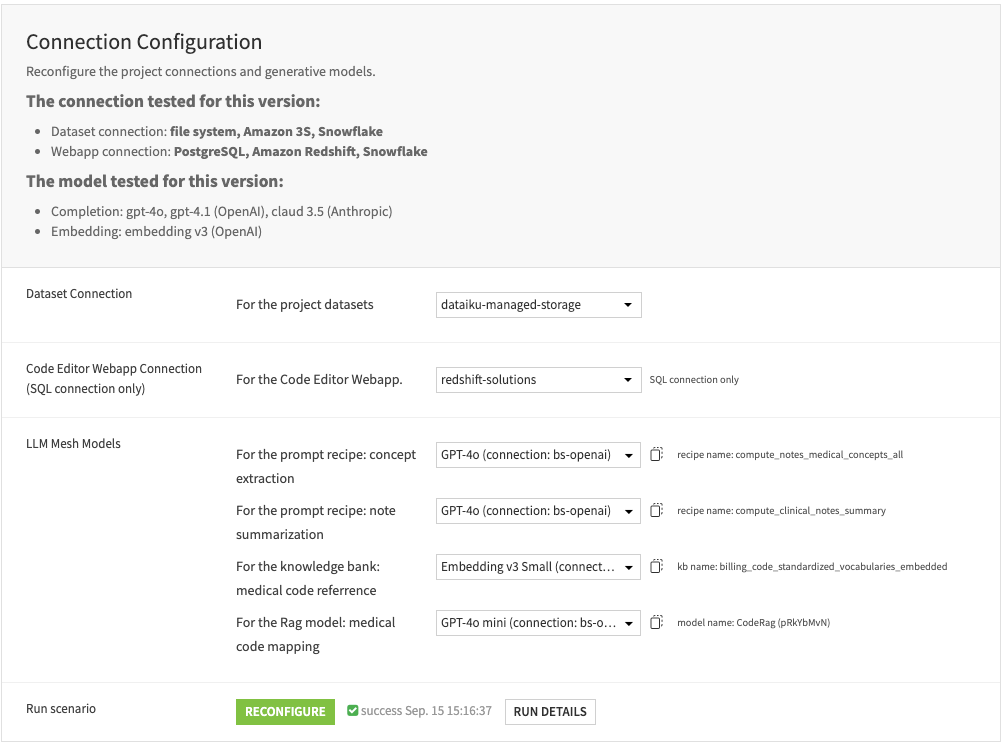
Connection configuration#
Within the Dataiku app, configure the connections for the project datasets and the Code Editor webapp. Select the generative models to power the LLM mesh recipes.
Tip
The Code Editor webapp requires an SQL connection to load the biomedical vocabularies. If a connection other than an SQL is selected, the webapp may not correctly display the “Mapped billing code” column with the linked record.
You can assign different generative models for each LLM Mesh recipe. Please refer to the Solution wiki chapter on “Dataiku Application” for more information.
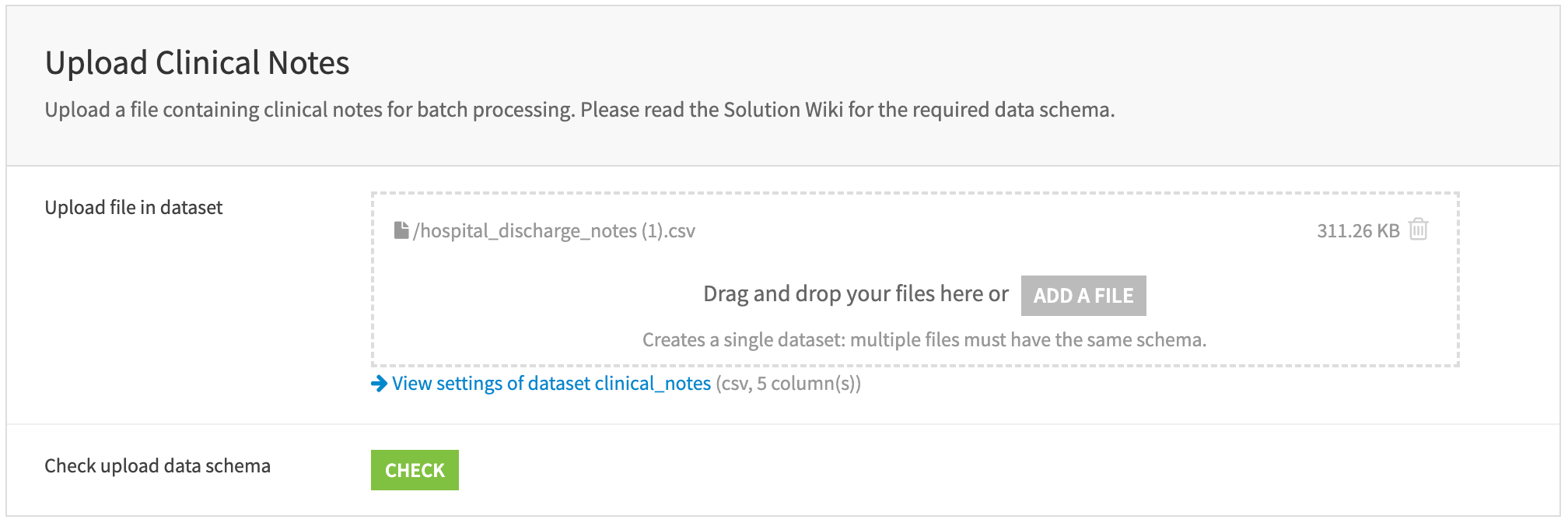
Upload clinical notes#
Upload a file containing clinical notes for batch processing. The file must include the following columns: patient_id, note_id, age, sex, and note. Each row should represent a single clinician’s note. Refer to the wiki chapter on Data Model to learn more about the required data schema.
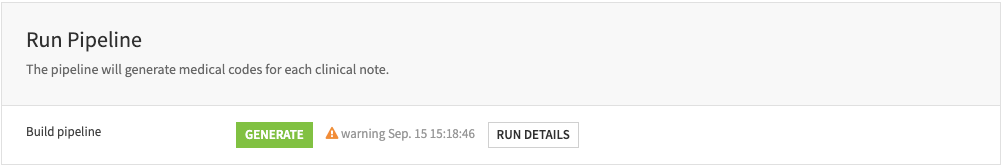
Run pipeline#
This step processes your configurations and ingests the clinical notes into the pipeline to generate medical billing codes. The pipeline includes the following steps:
Extract key clinical events and associated medical concepts from the clinical notes.
Generate a summary for each clinical note.
Embed the designed biomedical vocabularies—–including codes, labels, and definitions—–into a knowledge bank.
Assign the most relevant biomedical code from the knowledge bank to each medical concept.
Compute the model performance metrics.
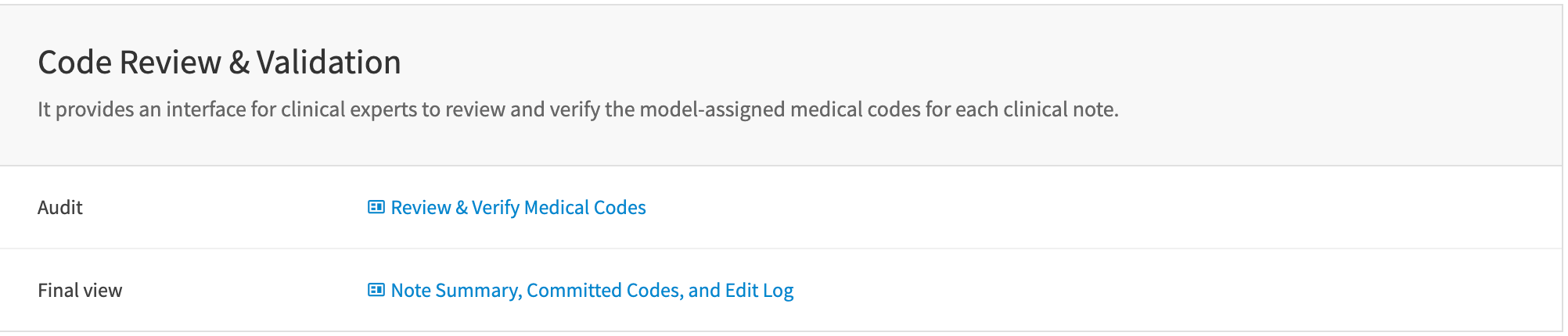
Launch the webapps#
Launch the webapps to review, modify, and verify the billing codes for each clinical note.
Create the model dashboard#
Launch the model metrics dashboard to review model performance.
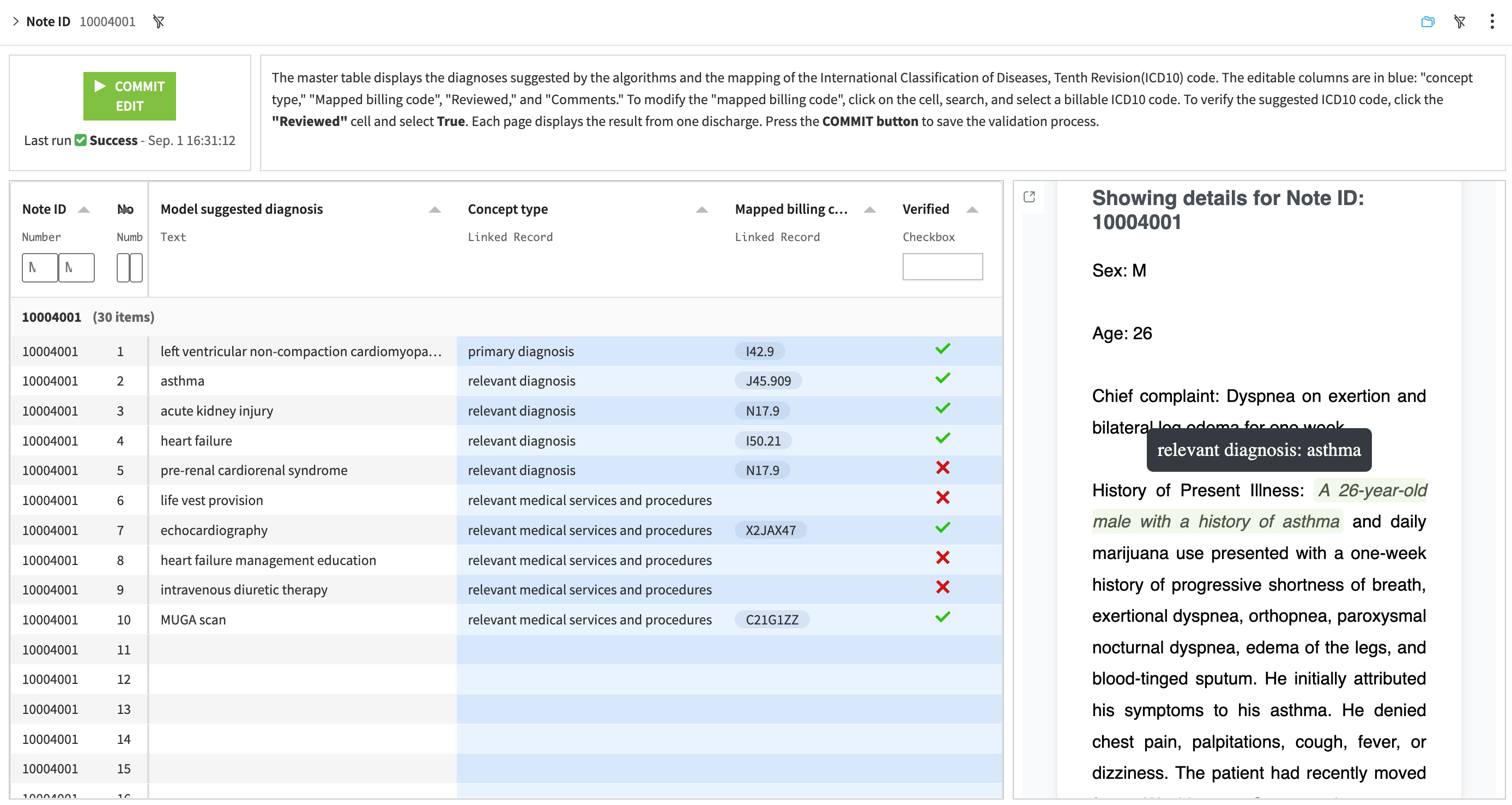
Webapp: Review & verify medical codes#
This webapp allows users to review, modify, and approve the model-assigned medical codes. This human-in-the-loop interface supports medical coders to verify the model-assigned codes for compliance with coding guidelines and regulations.
Select the note_id in the top left corner to review the model-assigned medical codes.
Modify the codes or concept domain and approve the results via the Code Editor webapp.
Review the original note with the associated clinical events.
Commit the approved medical codes by clicking on the top left button COMMIT EDIT .
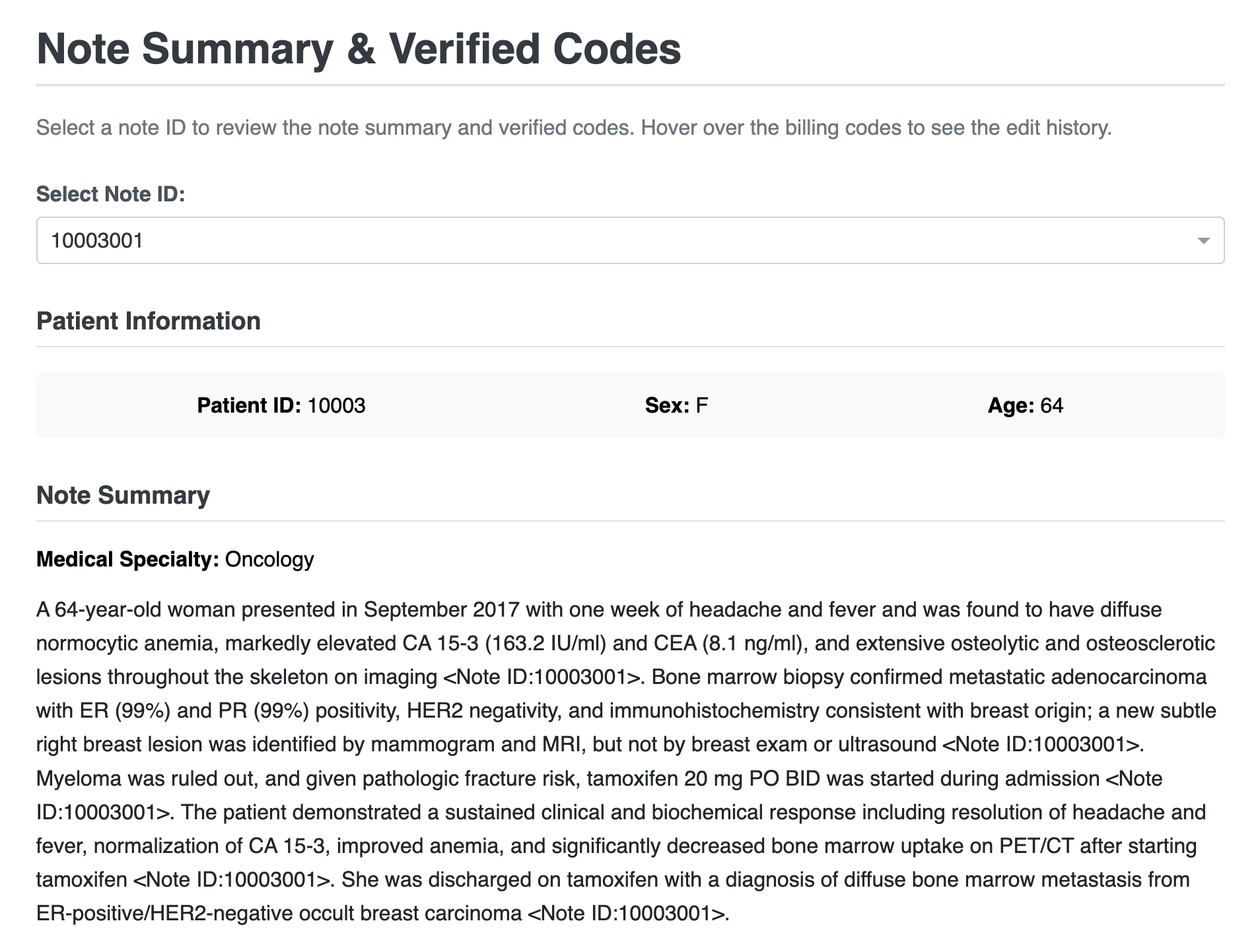
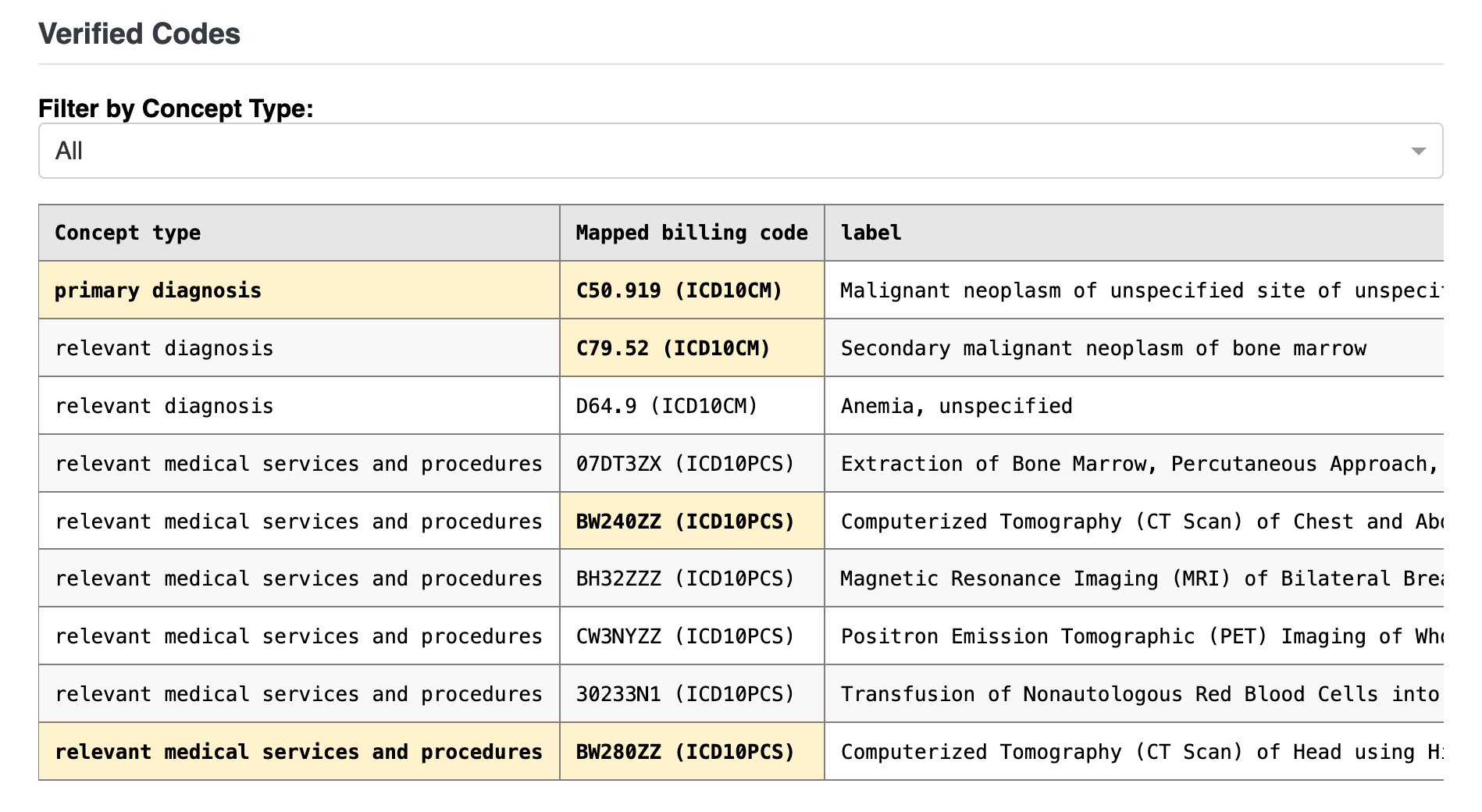
Webapp: Note summary, committed codes, and edit log#
This view facilitates clinicians authorizing the verified codes. It includes a model-generated summary and a complete edit history to track the changes made to the codes.
Dashboard: Model accuracy & code distribution#
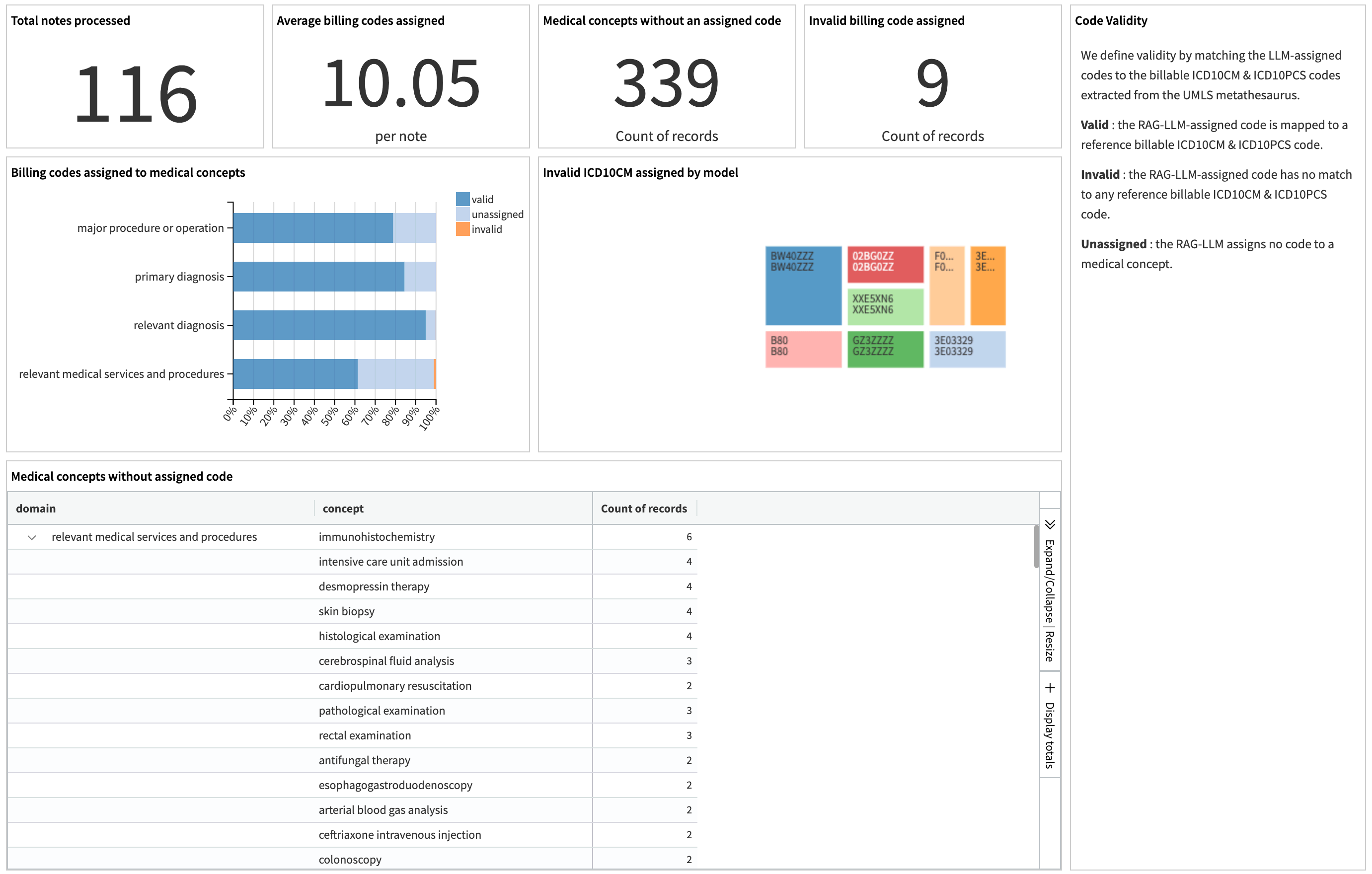
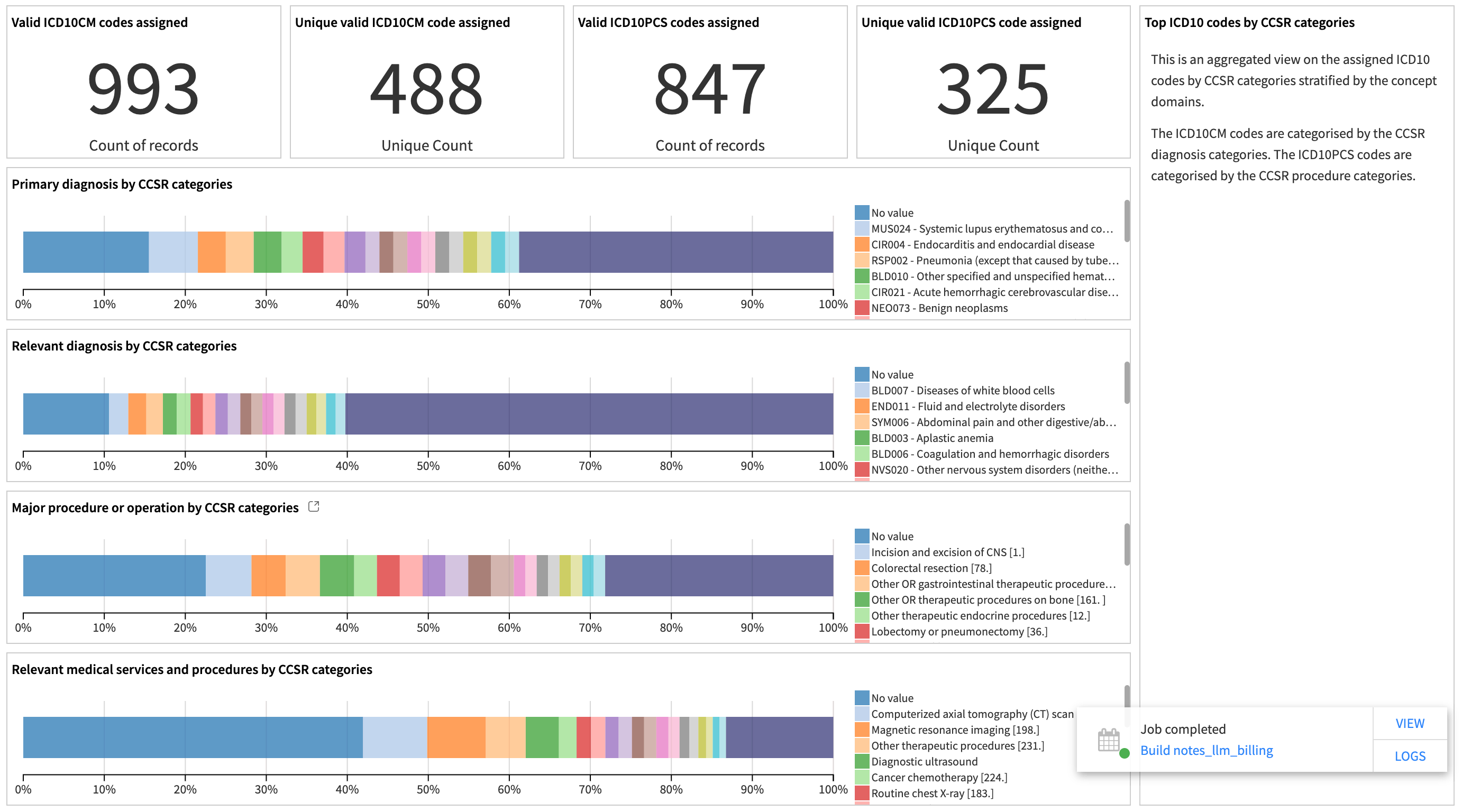
The dashboard displays a metrics panel for the data team to monitor model performance. It measures the quality of the generated medical codes by showing the ratio of valid, invalid, and missing codes among all extracted medical concepts. It also displays the distribution of the generated codes by diagnosis and procedure categories.
Model metrics#
These metrics describe the pipeline’s ability to extract critical medical events and map them to valid billing codes. They include the total number of clinical notes processed, the average codes assigned per note, code validity stratified by concept types, the count of unmapped medical concepts, and the model-generated invalid codes.
These plots show the top clinical categories of the model-assigned codes by different concept categories. They help the data team understand the data distribution and identify potential bias in the results.
Responsible AI statement#
In developing and deploying solutions like Medical Entity Extraction Assistant in healthcare, several responsible AI considerations should be addressed to ensure fairness, transparency, and accountability. Below are some key ethical considerations and potential biases concerning insight generation from observational health data from medical systems or longitudinal patient insurance claims.
Bias in input data#
Bias Type |
Description |
Recommendations |
|---|---|---|
Demographic bias |
If the input patient data over-represents specific demographics (for example age, gender, race, or location), it can lead to biased cohort insights. For example, if data skews toward urban areas, the Solution may not accurately capture the observational health outcomes in rural regions. |
Ensure data represents diverse patient social factors, health systems, and demographics. Regularly review and audit datasets created in cohorts to detect any demographic imbalances that could lead to biased or inaccurate insights derived from real-world patient data. |
Socioeconomic bias |
Data on patient populations may favor wealthier areas or practices due to inequity in healthcare access or social imbalances around seeking care or reimbursements for care, leading to bias against those serving lower-income communities. |
Balance datasets and evaluate patients in a cohort by including data from various economic and social factor strata and regions to ensure equitable representation. |
Data quality and source bias |
Input data may come from various sources (for example multiple claims, EMR systems, or syndicated data providers), each with its own biases and quality considerations. Potential duplicate patient records may also bias cohort incidence or prevalence estimates. |
Consider the limitations of each data source and use techniques like data augmentation, bias correction, quality metrics, and checks to ensure the quality of possibly disparate data sources doesn’t lead to biased patient population cohorts for further analysis. |
Health information generated by this Solution should promote unbiased and accurate insights from observational patient health signals and support research programs that improve patient outcomes, therapeutic interventions, and healthcare access—without reinforcing or exacerbating disparities or biases.
Caution and consideration of sample Solution data#
As a reminder, the synthetic data sources used in this Solution’s example application don’t reflect real distributions of patient or disease characterizations, and no insights or assumptions around observational health outcomes patterns should be made from the example insights derived from the patient cohorts. They shouldn’t be used in any further downstream business decision processes.
See also
We encourage users to check out Dataiku’s Responsible AI course to learn more.
Reproduce these processes with minimal effort#
This project equips healthcare and life science professionals to accelerate clinical operations by using Generative AI capabilities in Dataiku.
This Solution enables the healthcare organization to generate immediate insights from unstructured clinical text. Its framework can be modified and extended to fit your specific needs.
This documentation has provided several suggestions on how to derive value from this Solution. Ultimately however, the “best” approach will depend on your specific needs and data. If you’re interested in adapting this project to the specific goals and needs of your organization, Dataiku offers roll-out and customization services on demand.

